Background on knowledge representation and ontologies
As part of the ORION project, we have also made available two short introductory videos
1) Basics of ontologies and knowledge models (WATCH HERE)
2) The use of ontologies in health surveillance (WATCH HERE)
If you prefer a textual tutorial, read on below:
“An ontology defines a common vocabulary for researchers who need to share information in a domain. It includes machine-interpretable definitions of basic concepts in the domain and relations among them.”
(Noy and McGuinness, 2001)
Various initiatives have attempted to create a vocabulary for animal health data, such as SNOMED (Systematized Nomenclature of Medicine; http://www.snomed.org/snomed-ct\). Large networks of clinical practices will often adopt their own terminology for clinical recording. As in human medicine, insurance companies may also be particularly interested in setting standards for recording disease events.
Once we have a working vocabulary, why would we want to move beyond that, and build an ontology?
- “To share common understanding of the structure of information among people or software agents
- To enable reuse of domain knowledge
- To make domain assumptions explicit
- To separate domain knowledge from the operational knowledge
- To analyze domain knowledge”
(Noy and McGuinness, 2001).
To understand how ontologies provide these benefits, we will cover some theory focusing on the concepts highlighted above, in particular those two that are unique to ontologies: machine-interpretable definitions, and relationships.
Concepts and relationships
Think about standard vocabularies, or terminologies, as a list of concepts – _concepts _are the things we want to list or model, the things we want to represent in a specific domain.
We will work with 3 independent examples: A- a fictitious wine ontology, where we are interested in the concept “wine”; B- looking at a specific terminology, MESH (Medical Subject Headings), we will follow the concept “insulin”; C- still using MESH, we will follow the concept “Femoral Neck Fracture”.
Classes describe concepts in the domain. For example, a class of wines represents all wines. Specific wines are instances of this class. The Bordeaux wine in the glass in front of you while you read this document is an instance of the class of Bordeaux wines. A class can have subclasses that represent concepts that are more specific than the superclass. For example, we can divide the class of all wines into red, white, and rosé wines. Alternatively, we can divide a class of all wines into sparkling and nonsparkling (Noy and McGuinness, 2001).
This classes and subclasses structure is equivalent to the hierarchical structure of a vocabulary. Applying the logic to our other examples, “Hormone”, is a class, of which “Pancreatic Hormones” can be a subclass, and “Insulin” is, in turn, a sub-class of that. “Fracture” is a subclass of “Wounds and Injuries”.
In the wine example above, if we want to classify wines based on both suggested structures, we would have to combine all possible classes, resulting in: red sparkling and red non-sparkling wine; white sparkling and white non-sparkling and so on. As we add more dimensions, the list gets progressively complex. Adding new dimensions implies adding an exponentially growing number of concepts. Moreover, should we have red as a sub-class of wine, and sparkling as a sub-class of red, or the other way around? Try for instance finding all the sparkling wines in Figure 1A versus 1B.
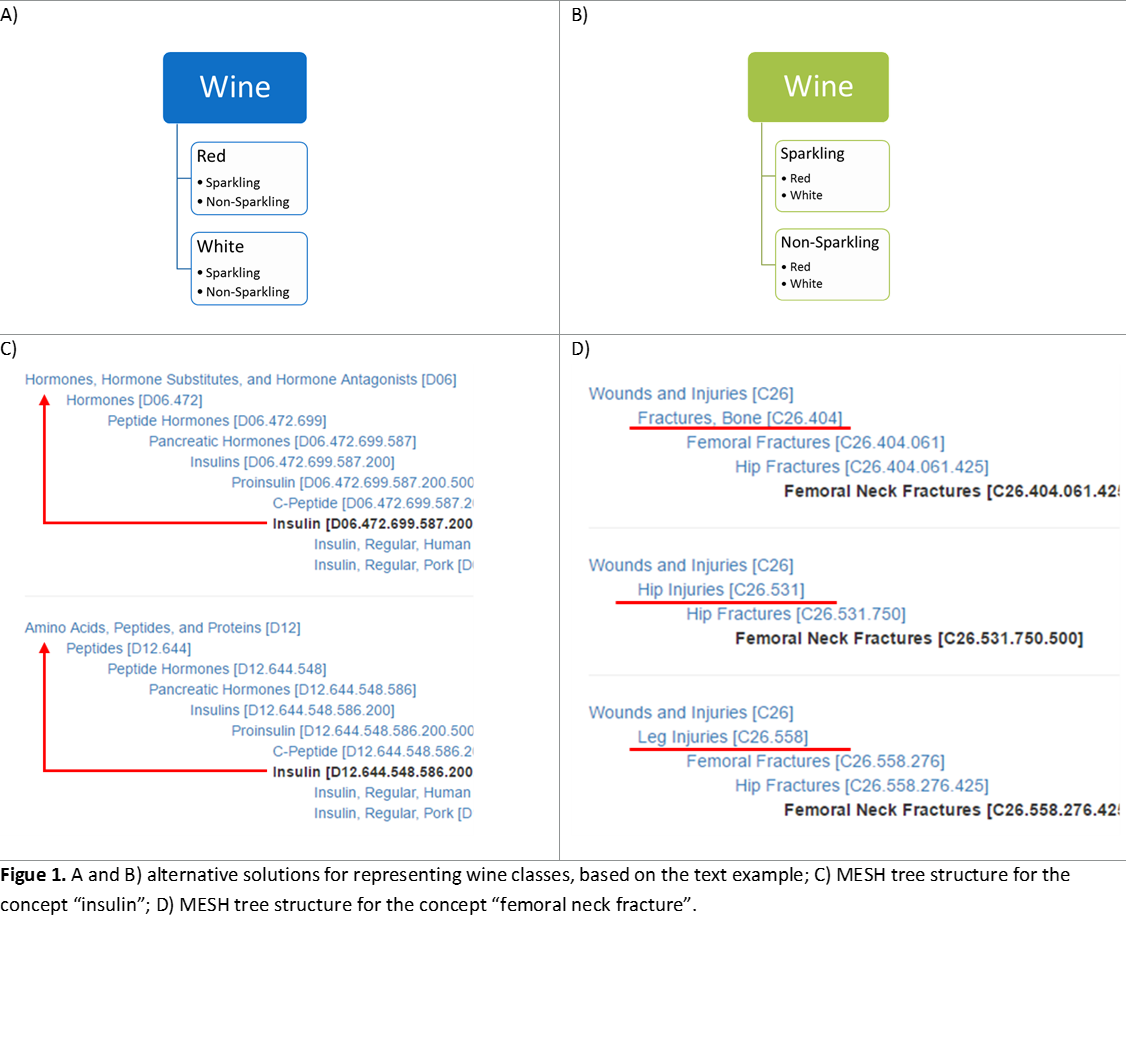
MESH dealt with the problem by entering the same concept in several different points of the vocabulary (a problem called “multiple inheritance”). Figure 1C and 1D show the tree structures for insulin and femoral neck fracture, respectively.
The complexity added to the lists comes from the complexity of the domains we are trying to model. “Red, White and Rosé” refer to the amount of pigmentation in the wine, while “sparkling or non-sparkling” refer to the presence of carbon dioxide bubbles. These are in reality different concepts, that could each be modelled using their own classes and subclasses, that is, each in their own hierarchical tree. Relationships (or properties) are then used to model the semantic connections between all the concepts. Compare figures 1A and 1B, now to Figure 2A.
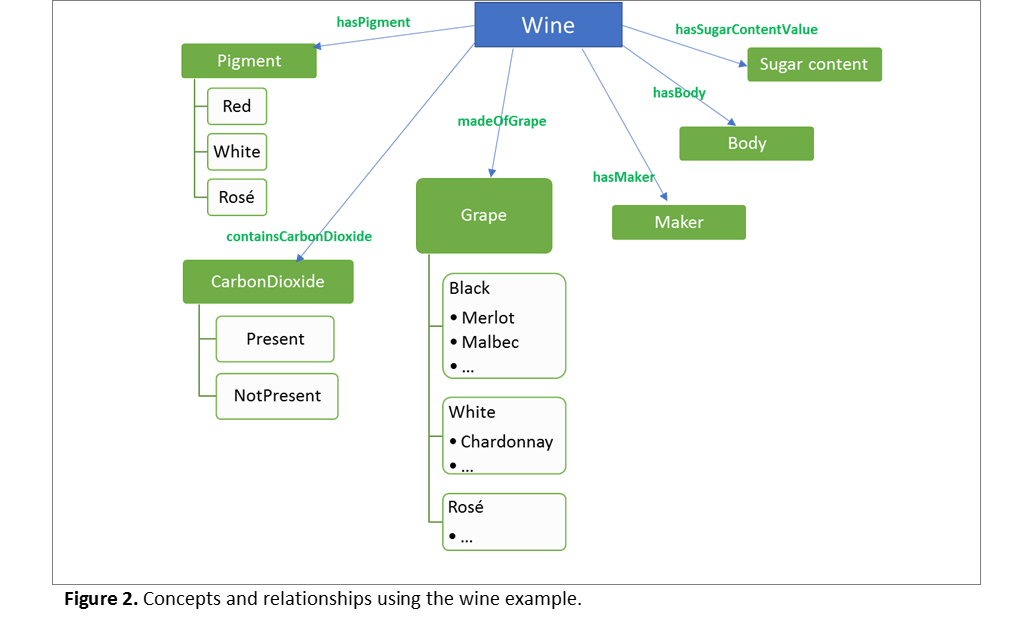
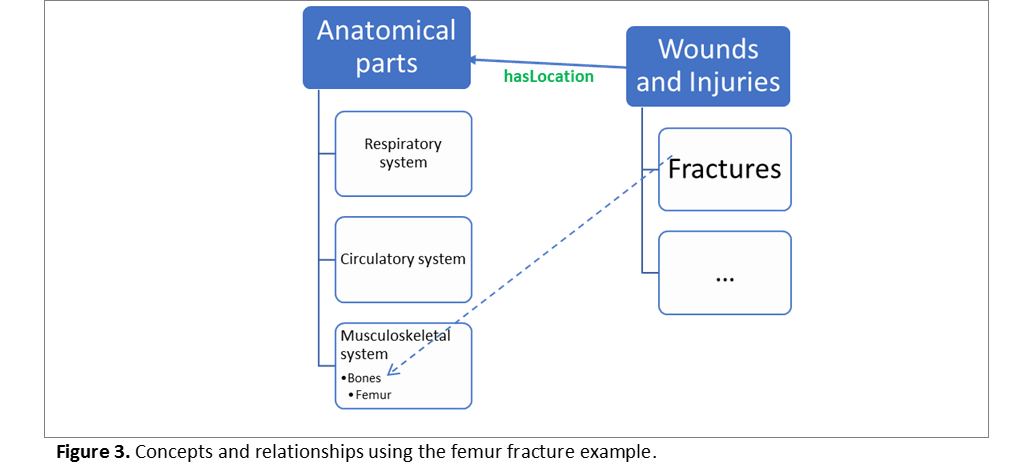
Similarly, Insulin as a molecule is a “Protein”, which has functional role “Hormone”. A Femur fracture is a bone injury, while the hip/leg tree structure shown in Figure 1D refers to the “location” of that fracture. Consider for instance the structure presented in Figure 3. There we have modelled (over-simplistically for the sake of this example): The hierarchical relationships among body parts, the hierarchical relationships among injuries, and then the overall property linking injuries and anatomical locations. In ontological language we can model these relationships in finer details, to specify for instance that “fracture is a type of injury that can only have as location a bone”.
 So far, we have only discussed the representation in “human readable format”. This form of knowledge representation on itself already presents some advantages in comparison to the use of a single hierarchical list of terms, such as making the knowledge structure explicit and facilitating the addition of concepts. It is also easier to evaluate, find and correct errors, and update the knowledge when the structure is made explicit like this.
So far, we have only discussed the representation in “human readable format”. This form of knowledge representation on itself already presents some advantages in comparison to the use of a single hierarchical list of terms, such as making the knowledge structure explicit and facilitating the addition of concepts. It is also easier to evaluate, find and correct errors, and update the knowledge when the structure is made explicit like this.
The full power of ontologies, however, comes from sharing a common understanding of the structure of information among people AND among machines.
MACHINE-INTERPRETABLE FORMATS: ONTOLOGY LANGUAGE
In the next section you can find additional materials to cover some background knowledge on the way we code ontologies (using the OWL language) and the way we represent data to support reasoning with ontologies (RDF framework). For the sake of this introductory material, we will just state that once a knowledge representation is agreed on, it will be coded into a language that makes it also understandable by the software applications in which it will be used.
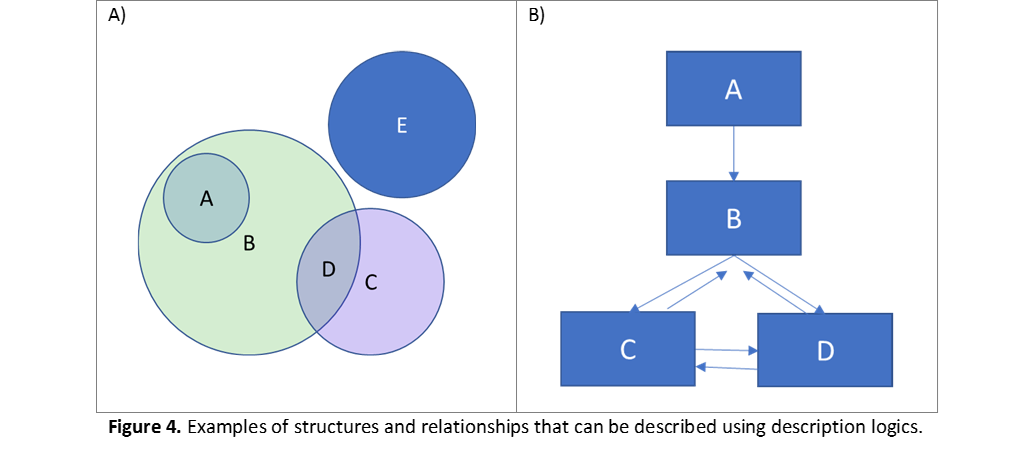
Say for instance that we are domain experts who agree on the knowledge representation shown in Figure 3 above. We, the domain experts, will draw the knowledge model, first defining the classes and their hierarchical structure; then defining the relationships among them. All details of our model will then be coded into the ontology. The language supports representation of these details using description logics. Consider the simple examples in Figure 4. Say that we, the experts, drew the relationships among the groups A-E. The ontology language supports representing this figure because it is capable of stating that A is contained in B (all elements in A are also elements of B, but not all elements in B belong to A); D is the result of the union between C and B; and E is disjoint from all other classes. Relationships can also have a number of properties. Consider now Figure 4B. Having the knowledge that this is a genealogical tree, we as modelers would have enforced rules such as: if B is mother of C, then C is child of B; a mother (say B) can have any number of children, but a child can only have one mother (cardinality rules); if A is an ancestor of B, then it is also an ancestor of C and D (transitive rules); if C is sibling to D, then D is sibling to C (reflective rule). A language powered with description logics will translate all of these rules into machine-interpretable format.
 Why is this important?
Why is this important?
Machine-interpretable version of a knowledge model (ontology) allows:
1) Using reasoners. Reasoners are software capable of inferring logical consequences based on the semantic (relationships) knowledge coded into the ontology. In the example of Figure 3, for instance, once we code that fractures are injuries which can only happen in bones, the reasoner could be used to detect errors (for instance if someone tries to declare a fracture in a location that is not a bone) or infer things, such that femur is a bone. This kind of inference and error checking gets quickly out of hand for humans to do once a knowledge base grows. Making the semantic structure and assumptions explicit, in machine-readable format, allows us to employ software to do it.
2) Knowledge growth and updates. Growing and updating knowledge is easier in a format where structure and relationships are explicit. Growing: automated and semi-automated methods can be used to learn new classes from data. Updating: say for instance that the name of the bone “femur” changed, or even a new bone is found in the body. Updating the ontology would require only adding the new bone to the class of bones, rather than including new codes for all possible injuries related to that bone.
3) Reuse. Ontologies are meant to model specific pieces of knowledge, in a way that allows linking to complementary pieces. Once again look at Figure 3. When building a model like that, we could just import (reuse) an already existing ontology of anatomical parts. Ontologies can be reused on the whole, or single concepts (classes) can be imported and reused. We can for instance reuse an entire ontology of infectious diseases, or just import the concept “brucellosis”. We can import the hierarchical structure, or also the relationships associated with the concept: which pathogens are associated with the disease, which host species are susceptible, which clinical signs are associated with it, etc.
4) Distributed knowledge. Related to the idea of reusing, ontologies allow for knowledge to be distributed over many sources. Consider for instance the diagram in Figure 4B. Say one ontology has coded the knowledge that B is mother of C. If we reuse that knowledge but add that A is mother of B, the reasoner can infer that A is grandmother to C. We therefore build on existing knowledge. The same principle can be used to query distributed data (from multiple sources) using the ontology. However, in the context of the animal health surveillance ontology, we are assuming that data will continue to be private, and we will discuss only sharing a public ontology, NOT sharing data. The important thing to mention is that the use of a common ontology allows us not only to share knowledge with others, but to build that knowledge base together, with different pieces of knowledge coming from different sources.
5) Interoperability. Vocabularies allow humans to understand each other and agree on what things mean. Ontologies allow software to talk to each other. This interoperability means that we would be able to share tools. Developing tools to analyse data then also becomes a community project – any tools developed by one team can be readily employed by another group to analyse their own data. Even if data are coded using different practices and standards, applying the ontology then means that everything gets “translated” into a common language, and can be then analysed using the same tools, with compatible/comparable results. The data themselves continue to be private.
Reference
Natalya F. Noy and Deborah L. Mcguinness. 2001. Ontology Development 101: A Guide to Creating Your First Ontology. Available at http://protege.stanford.edu/publications/ontology_development/ontology101.pdf.